Research
We use experimental and computational tools to understand the functional effects of amino acid mutations, the biophysical origins of these effects, and their impacts on evolution. We seek to understand basic aspects of protein evolution while addressing key questions in virology and immunology, such as the evolution of SARS-CoV-2 and related animal viruses, and the somatic development of broadly protective antibodies.
Areas of Research
-

Deep Mutational Scanning
We use “deep mutational scanning” methods to precisely measure the effects of mutations on protein function in high throughput.
-

SARS-CoV-2 Evolution
The SARS-CoV-2 pandemic has been marked by the emergence of viral variants with mutations that impact transmission and immune recognition.
-

Zoonotic Virus Evolution
Animal viruses evolve latent traits that enable human infection and pandemic spillover.
-

Antibody Evolution
To counteract viral evolution, our bodies deploy an arsenal of protective antibodies that evolve via the somatic process of affinity maturation.
-

Epistasis in Protein Evolution
Epistasis—the non-additivity of mutational effects—shapes the trajectories available to an evolving protein and highlights peculiarities in protein folding and function.
Deep Mutational Scanning
We use “deep mutational scanning” methods to precisely measure the effects of mutations on protein function in high throughput. These methods allow us to determine the impacts of many single mutations or combinations of mutations on key protein properties such as affinity for ligand or folding efficiency. The resulting maps elucidate the relationships between protein sequence, structure, and function, and highlight the molecular features that shape subsequent protein evolution.
SARS-CoV-2 Evolution
The SARS-CoV-2 pandemic has been marked by the emergence of viral variants with mutations that impact transmission and immune recognition. We have developed platforms to prospectively study the effects of mutations in key domains of the SARS-CoV-2 spike on phenotypes including ACE2 receptor binding affinity, protein folding stability, and recognition by antiviral antibodies (including those used in the therapeutic treatment of COVID-19). This work aids in the continued surveillance and modeling of viral evolution and informs the development of the next generation of antibody and vaccine therapeutics.
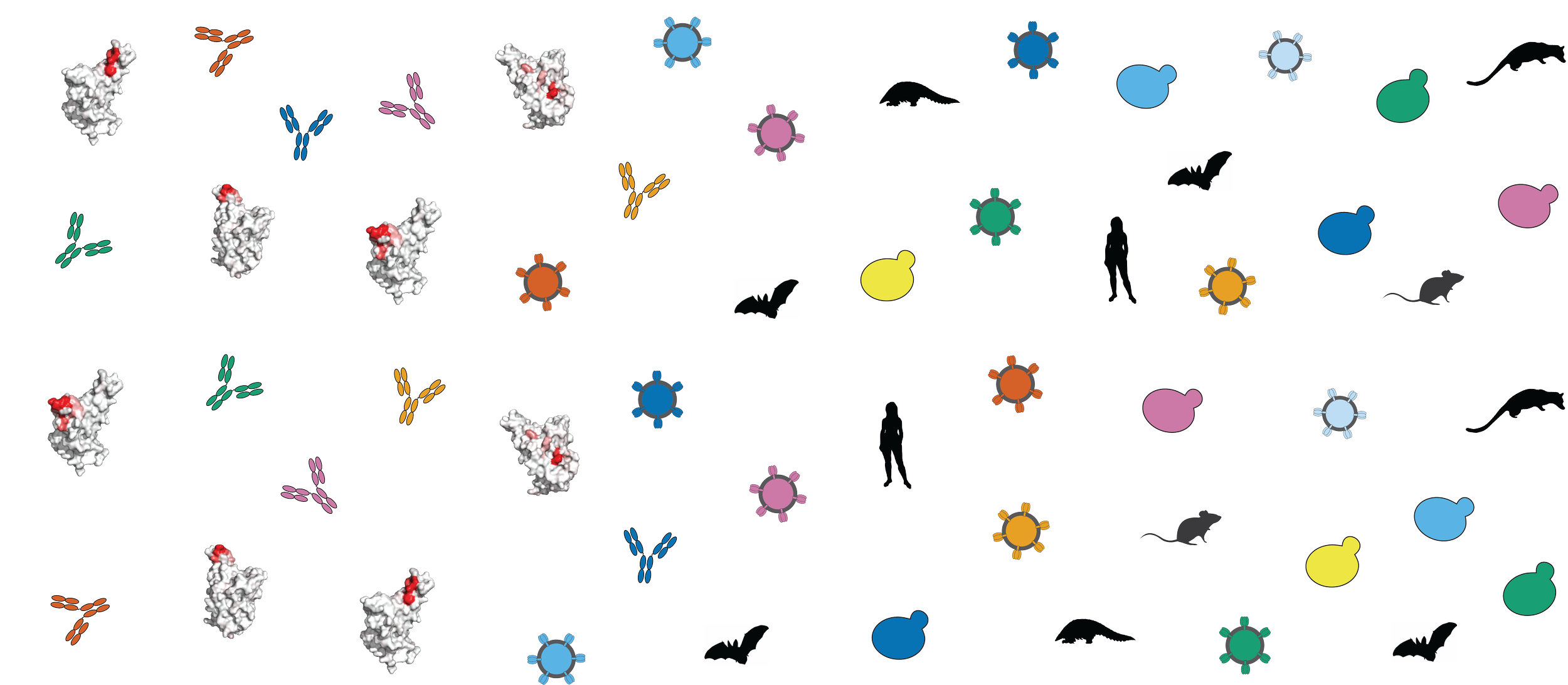
Zoonotic Virus Evolution
Animal viruses evolve latent traits that enable human infection and pandemic spillover. For example, SARS-related coronaviruses circulate in Rhinolophus bat reservoirs where they acquired the ability to interact with ACE2 receptors from humans and other intermediate hosts. We seek to understand the molecular evolutionary, immunological, and ecological factors that drive shifts in receptor specificities in animal viruses. This work enables enhanced surveillance and pandemic preparedness against viral spillovers from diverse and potentially unknown viral sub-lineages.
Antibody Evolution
To counteract viral evolution, our bodies deploy an arsenal of protective antibodies that evolve via the somatic process of affinity maturation. We seek to understand how mutations impact the dynamics of antibody maturation and yield antibodies with improved potency and breadth of viral recognition. These efforts are key to the development of broad therapeutics against diverse endemic viruses (e.g., HIV) or viral families of future pandemic potential.
Epistasis in Protein Evolution
Epistasis—the non-additivity of mutational effects—shapes the trajectories available to an evolving protein and highlights peculiarities in protein folding and function. We are interested in the prevalence and mechanisms by which mutations combine idiosyncratically and how epistasis influences the topology of fitness landscapes.




